class: center, middle, inverse, title-slide # Multiple Regression III --- ## Last time... * Model comparison * Categorical predictors/dummy coding -- ### Today... *ANOVA (the 611 version) It's important to note that by covering dummy coding a categorical variable, we have already covered ANOVA -- there is nothing more you can learn from this technique that the omnibus test of that model will not tell you. --- ## Example Participants performed an eye-hand coordination task while subjected to periodic 3-second bursts of 85 dB white noise played over earphones. The task required participants to keep a mouse pointer on a red dot that moved in a circular motion at a rate of 1 revolution per second. Participants performed the task until they allowed the pointer to stray from the rotating dot 10 times. The time (in seconds) at the 10th failure was recorded and is the outcome measure.  --- ### Example The participants were randomly assigned to one of four noise conditions: * controllable and predictable noise * uncontrollable but predictable noise * controllable but unpredictable noise * uncontrollable and unpredictable noise. When noise was predictable, the 3-second bursts of noise would occur regularly every 20 seconds. When noise was unpredictable, the 3-second bursts would occur randomly (although every 20 seconds on average). --- When noise was uncontrollable, participants could do nothing to prevent the noise from occurring. When noise was controllable, participants were shown a button that would prevent the noise, but they were told, "the button is a safety measure, for your protection, but we would prefer that you not use it unless absolutely necessary." No participants actually used the button. Why is it important that the button was never used? Why is random assignment important in this design? --- ```r library(here) rotate = read.csv(here("data/pursuit_rotor.csv")) head(rotate) ``` ``` ## ID Predict Control Group Time ## 1 1 0 1 3 504 ## 2 2 0 0 1 443 ## 3 3 1 1 4 292 ## 4 4 0 1 3 398 ## 5 5 1 1 4 119 ## 6 6 0 0 1 545 ``` ```r class(rotate$Group) ``` ``` ## [1] "integer" ``` ```r rotate$Group_lab = factor(rotate$Group, labels = c("Controllable\nPredictable", "Uncontrollable\nPredictable", "Controllable\nUnpredictable", "Uncontrollable\nUnpredictable")) ``` --- ```r library(ggpubr) ggboxplot(data = rotate, x = "Group_lab", y = "Time", xlab = F) ``` 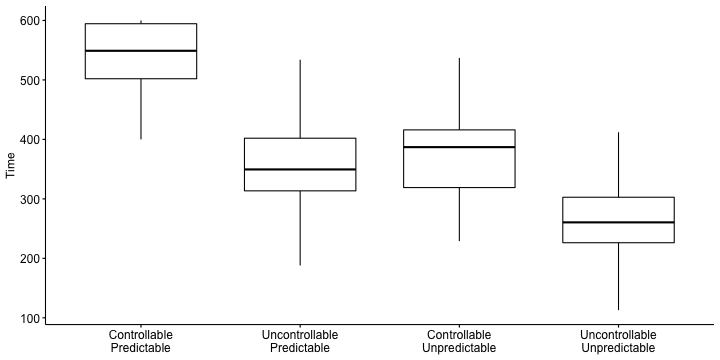<!-- --> -- The pattern of means indicates that performance degrades with either uncontrollable noise or unpredictable noise. Noise that is both uncontrollable and unpredictable appears to be particularly disruptive. --- ```r ggbarplot(data = rotate, x = "Group_lab", y = "Time", xlab = F, add = c("mean_ci"), fill = "purple") ``` 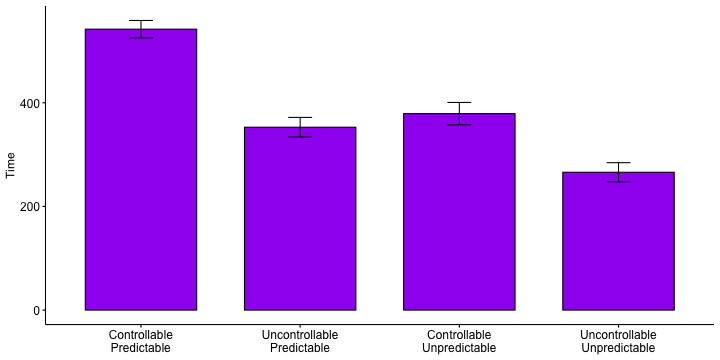<!-- --> Addition of the confidence intervals indicates that the two extreme conditions are likely different from all of the other conditions. --- ```r library(knitr) library(kableExtra) rotate %>% group_by(Group_lab) %>% summarize(N = n(), Mean = mean(Time, na.rm = T), SD = sd(Time, na.rm = T)) %>% kable(., digits = 2) %>% kable_styling() ``` <table class="table" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Group_lab </th> <th style="text-align:right;"> N </th> <th style="text-align:right;"> Mean </th> <th style="text-align:right;"> SD </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> Controllable Predictable </td> <td style="text-align:right;"> 47 </td> <td style="text-align:right;"> 542.04 </td> <td style="text-align:right;"> 57.11 </td> </tr> <tr> <td style="text-align:left;"> Uncontrollable Predictable </td> <td style="text-align:right;"> 52 </td> <td style="text-align:right;"> 352.85 </td> <td style="text-align:right;"> 67.98 </td> </tr> <tr> <td style="text-align:left;"> Controllable Unpredictable </td> <td style="text-align:right;"> 45 </td> <td style="text-align:right;"> 379.04 </td> <td style="text-align:right;"> 71.85 </td> </tr> <tr> <td style="text-align:left;"> Uncontrollable Unpredictable </td> <td style="text-align:right;"> 56 </td> <td style="text-align:right;"> 265.98 </td> <td style="text-align:right;"> 68.68 </td> </tr> </tbody> </table> The groups differ in their sample sizes, which can easily occur with free random assignment. There are advantages to equal sample sizes, so researchers often restrict random assignment to insure equal sample sizes across conditions. Hard to tell if the variability in the four groups is homogeneous. --- ### Hypothesis Unlike regression, the ANOVA framework has a single hypothesis test. This is equivalent to the omnibus test of a multiple regression model. .pull-left[ ### Regression `$$H_0: \rho^2_{Y\hat{Y}} = 0$$` `$$H_1: \rho^2_{Y\hat{Y}} > 0$$` ] .pull-left[ ### ANOVA `$$H_0: \mu_1 = \mu_2 = \mu_3 = \mu_4$$` `$$H_1: \text{Not that } \mu_1 = \mu_2 = \mu_3 = \mu_4$$` This can occur in quite a number of ways. ] --- The total variability of all of the data, regardless of group membership, can be expressed as: `$$\large Var(Y) = \frac{1}{N}\sum_{k=1}^G\sum_{i=1}^{N_k}(Y_{ik}-\bar{Y})^2$$` for G groups and `\(N_k\)` participants within groups. In analysis of variance, we will be interested in the numerator of this variance equation, known as the total sum of squares: `$$\large SS_{tot} = \sum_{k=1}^G\sum_{i=1}^{N_k}(Y_{ik}-\bar{Y})^2$$` It's worth noting that this is just a more complicated way of expressing total sums of squares, but in a way that will be useful for thinking about how we partition sums of squares later. --- We already know from regression that the deviation of a score from the grand mean is the sum of two independent parts. In regression these parts are the deviation of the actual score from the predicted score, and the deviation of the predicted value from the grand mean. `$$\large Y_{i}-\bar{Y} = (Y_{i}-\hat{Y}_i) + (\hat{Y} - \bar{Y}_i)$$` In ANOVA, this holds true, but we express these relationships by referring to each Y within a group, and instead of "predicted value" we talk about group means. So now the parts are the deviation of the score from its group mean, and the deviation of that group mean from the grand mean. Why do we substitute "group mean" for "predicted value"? `$$\large Y_{ik}-\bar{Y} = (Y_{ik}-\bar{Y}_k) + (\bar{Y}_k - \bar{Y})$$` -- In other words, each deviation score has a within-group part and a between-group part. These separate parts can be squared and summed, giving rise to two other sums of squares. --- One part, the within-groups sum of squares, represents squared deviations of scores around the group means: `$$\large SS_w = \sum_{k=1}^G\sum_{i=1}^{N_k}(Y_{ik}-\bar{Y}_k)^2$$` The other, the between-groups sum of squares, represents deviations of the group means around the grand mean: `$$\large SS_b = \sum_{k=1}^GN_k(\bar{Y}_{k}-\bar{Y})^2$$` How do these compare the sums of squares we are familiar with in regression? --- Sums of squares are central to ANOVA. They are the building blocks that represent different sources of variability in a research design. They are additive, meaning that the `\(SS_{tot}\)` can be partitioned, or divided, into independent parts. The total sum of squares is the sum of the between-groups sum of squares and the within-groups sum of squares. `$$\large SS_{tot} = SS_b + SS_w$$` The relative magnitude of sums of squares, especially in more complex designs, provides a way of identifying particularly large and important sources of variability. --- If the null hypothesis is true, then variability among the means should be consistent with the variability of the data. We know that relationship already: `$$\large \hat{\sigma}^2_{\bar{Y}} = \frac{\hat{\sigma}^2_{Y}}{N}$$` In other words, if we have an estimate of the variance of the means, we can transform that into an estimate of the variance of the scores, provided the only source of mean variability is sampling variability (the null hypothesis). `$$\large N\hat{\sigma}^2_{\bar{Y}} = \hat{\sigma}^2_Y$$` --- The `\(SS_w\)` is qualitatively giving us information that is similar to `$$\large \hat{\sigma}^2_Y$$` The `\(SS_b\)` is qualitatively giving us information that is similar to `$$\large N\hat{\sigma}^2_{\bar{Y}}$$` These are arrived at separately, but under the null hypothesis they should be estimates of the same thing. The only reason that `\(SS_b\)` would be larger than expected is if there are systematic differences among the mean, perhaps created by experimental manipulations. We need a formal way to make the comparison. --- Because the sums of squares are numerators of variance estimates, we can divide each by their respective degrees of freedom to get variance estimates that, under the null hypothesis, should be approximately the same. These variance estimates are known as mean squares: .pull-left[ `$$\large MS_w = \frac{SS_w}{df_w}$$` `$$\large df_w = N-G$$` ] .pull-right[ `$$\large MS_b = \frac{SS_b}{df_b}$$` `$$\large df_b = G-1$$` ] How are these degrees of freedom determined? --- ANOVA assumes homogeneity of group variances. Under that assumption, the separate group variances are pooled to provide a single estimate of within-group variability. `$$\large SS_w = \sum_{k=1}^G\sum_{i=1}^{N_k}(Y_{ik}-\bar{Y}_k)^2 = \sum_{k=1}^G(N_k-1)\hat{\sigma}^2_k$$` The degrees of freedom are likewise pooled: `$$\large df_w = N-G=(N_1-1)+(N_2-1)+\dots+(N_G-1)$$` Where have we seen similar pooling? --- If data are normally distributed, then the variance is `\(\chi^2\)` distributed. The ratio of two `\(\chi^2\)` distributed variables is F distributed. In analysis of variance, the ratio of the mean squares (variance estimates) is symbolized by `\(F\)` and has a sampling distribution that is `\(F\)` distributed with degrees of freedom equal to `\(df_b\)` and `\(df_w\)`. That sampling distribution is used to determine if an obtained `\(F\)` statistic is unusual enough to reject the null hypothesis. `$$\large F = \frac{MS_b}{MS_w}$$` --- `$$\large F = \frac{MS_b}{MS_w}$$` If the null hypothesis is true, `\(F\)` has an expected value of approximately 1.00 (the numerator and denominator are estimates of the same variability). If the null hypothesis is false, the numerator will be larger than the denominator because systematic, between-group differences contribute to the variance of the means, but not to the variance within groups (ideally). If `\(F\)` is "large enough," we will reject the null hypothesis as a reasonable description of the obtained variability. --- The `\(F\)` statistic can be viewed as follows: `$$\large F = \frac{\hat{Q} + \hat{\sigma}^2}{\hat{\sigma}^2}$$` The extra term in the numerator (Q) is the treatment effect that, if present, increases variability among the means but does not affect the variability of scores within a group (treatment is a constant within any group). As the treatment effect gets larger, the `\(F\)` statistic departs from 1.00. If it departs enough, we claim F to be rare or unusual under the null and reject the null. The `\(F\)` probability density distribution tells us how rare or unusual a particular `\(F\)` statistic happens to be. The shape of the `\(F\)` distribution is defined by its parameters, `\(df_b\)` and `\(df_w\)`. --- 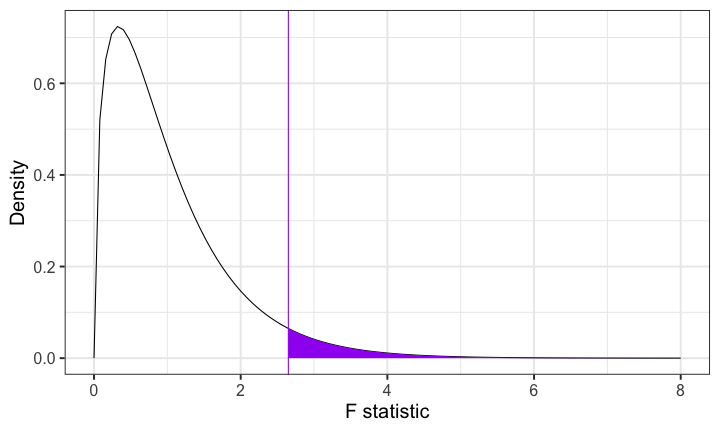<!-- --> For 3 and 196 degrees of freedom, an `\(F\)` statistic of 2.65 or greater will indicate between-groups variability relative to within-group variability that is unusual if the null hypothesis is true. --- ANOVA for the sample data: ```r fit_1 = aov(Time ~ Group_lab, data = rotate) summary(fit_1) ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## Group_lab 3 2000252 666751 149.8 <2e-16 *** ## Residuals 196 872288 4450 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` The `\(F\)` statistic is highly unusual in the `\(F\)` distribution, assuming the null hypothesis is true. We reject the null hypothesis. Note: how does the output above compare to this output? What is the difference between the two? ```r fit_1.1 = aov(Time ~ Group, data = rotate) summary(fit_1.1) ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## Group 1 1613798 1613798 253.9 <2e-16 *** ## Residuals 198 1258741 6357 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` --- ```r fit_1 = aov(Time ~ Group_lab, data = rotate) summary(fit_1) ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## Group_lab 3 2000252 666751 149.8 <2e-16 *** ## Residuals 196 872288 4450 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` We know the means are not equal, but the particular source of the inequality is not revealed by the `\(F\)` test. One simple way to determine what is behind the significant `\(F\)` test is to compare each condition to all other conditions. These pairwise comparisons can quickly grow in number as the number of conditions increases. With 4 (k) conditions, we have k(k-1)/2 = 6 possible pairwise comparisons. --- As the number of groups in the ANOVA grows, the number of possible pairwise comparisons increases dramatically. <!-- --> --- As the number of significance tests grows, and assuming the null hypothesis is true, the probability that we will make one or more Type I errors increases. To approximate the magnitude of the problem, we can assume that the multiple pairwise comparisons are independent. The probability that we don’t make a Type I error for just one test is: `$$\large P(\text{No Type I}, 1 \text{ test}) = 1-\alpha$$` The probability that we don't make a Type I error for two tests is (note the reason we assume independence): `$$\large P(\text{No Type I}, 2 \text{ test}) = (1-\alpha)(1-\alpha)$$` --- For C tests, the probability that we make **no** Type I errors is `$$\large P(\text{No Type I}, C \text{ tests}) = (1-\alpha)^C$$` We can then use the following to calculate the probability that we make one or more Type I errors in a collection of C independent tests. `$$\large P(\text{At least one Type I}, C \text{ tests}) = 1-(1-\alpha)^C$$` As the number of comparisons grows, the inflation of the Type I error rate can be substantial. --- The Type I error inflation that accompanies multiple comparisons motivates the large number of "correction" procedures that have been developed. <!-- --> --- Multiple comparisons, each tested with `\(\alpha_{per-test}\)`, increases the family-wise `\(\alpha\)` level. `$$\large \alpha_{family-wise} = 1 - (1-\alpha_{per-test})^C$$` Šidák showed that the family-wise a could be controlled to a desired level (e.g., .05) by changing the `\(\alpha_{per-test}\)` to: `$$\large \alpha_{per-wise} = 1 - (1-\alpha_{family-wise})^{\frac{1}{C}}$$` Bonferroni (and Dunn, and others) suggested this simple approximation: `$$\large \alpha_{per-test} = \frac{\alpha_{family-wise}}{C}$$` This is typically called the Bonferroni correction and is very often used even though better alternatives are available. --- ```r pc_1 = pairwise.t.test(x = rotate$Time, g = rotate$Group, p.adjust.method = "none") pc_1 ``` ``` ## ## Pairwise comparisons using t tests with pooled SD ## ## data: rotate$Time and rotate$Group ## ## 1 2 3 ## 2 < 2e-16 - - ## 3 < 2e-16 0.055 - ## 4 < 2e-16 1.5e-10 5.9e-15 ## ## P value adjustment method: none ``` ```r pc_2 = pairwise.t.test(x = rotate$Time, g = rotate$Group, p.adjust.method = "bonferroni") pc_2 ``` ``` ## ## Pairwise comparisons using t tests with pooled SD ## ## data: rotate$Time and rotate$Group ## ## 1 2 3 ## 2 < 2e-16 - - ## 3 < 2e-16 0.33 - ## 4 < 2e-16 9.2e-10 3.6e-14 ## ## P value adjustment method: bonferroni ``` --- The Bonferroni procedure is conservative. Other correction procedures have been developed that control family-wise Type I error at .05 but that are more powerful than the Bonferroni procedure. The most common one is the Holm procedure. The Holm procedure does not make a constant adjustment to each per-test `\(\alpha\)`. Instead it makes adjustments in stages depending on the relative size of each pairwise p-value. 1. Rank order the p-values from largest to smallest. 2. Start with the smallest p-value. Multiply it by its rank. 3. Go to the next smallest p-value. Multiply it by its rank. If the result is larger than the previous step, keep it. Otherwise replace with the previous step adjusted p-value. 4. Repeat Step 3 for the remaining p-values. 5. Judge significance of each new p-value against `\(\alpha = .05\)`. --- <table class="table" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:right;"> Original p value </th> <th style="text-align:right;"> Rank </th> <th style="text-align:right;"> Rank x p </th> <th style="text-align:right;"> Holm </th> </tr> </thead> <tbody> <tr> <td style="text-align:right;"> 0.0023 </td> <td style="text-align:right;"> 6 </td> <td style="text-align:right;"> 0.0138 </td> <td style="text-align:right;"> 0.0138 </td> </tr> <tr> <td style="text-align:right;"> 0.0012 </td> <td style="text-align:right;"> 5 </td> <td style="text-align:right;"> 0.0060 </td> <td style="text-align:right;"> 0.0138 </td> </tr> <tr> <td style="text-align:right;"> 0.0450 </td> <td style="text-align:right;"> 4 </td> <td style="text-align:right;"> 0.1800 </td> <td style="text-align:right;"> 0.1800 </td> </tr> <tr> <td style="text-align:right;"> 0.0470 </td> <td style="text-align:right;"> 3 </td> <td style="text-align:right;"> 0.1410 </td> <td style="text-align:right;"> 0.1800 </td> </tr> <tr> <td style="text-align:right;"> 0.0530 </td> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 0.1060 </td> <td style="text-align:right;"> 0.1800 </td> </tr> <tr> <td style="text-align:right;"> 0.2100 </td> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 0.2100 </td> <td style="text-align:right;"> 0.2100 </td> </tr> </tbody> </table> A single adjusted per-test a would be used with the Bonferroni procedure: .05/6 = .0083. We would compare each original p-value to this new criterion and claim as significant only those that are less. Or, we could multiply the original p-values by 6 and judge them against a =.05. --- Performing the test using the `lm()` function means that some of the possible pairwise comparisons are built in: ```r rotate$Group = as.factor(rotate$Group) coef(summary(lm(Time~Group, data = rotate))) ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 542.0426 9.730897 55.70325 4.073456e-122 ## Group2 -189.1964 13.426685 -14.09107 1.331759e-31 ## Group3 -162.9981 13.913633 -11.71499 2.267662e-24 ## Group4 -276.0604 13.197069 -20.91831 7.933911e-52 ``` ```r pairwise.t.test(x = rotate$Time, g = rotate$Group, p.adjust.method = "holm") ``` ``` ## ## Pairwise comparisons using t tests with pooled SD ## ## data: rotate$Time and rotate$Group ## ## 1 2 3 ## 2 < 2e-16 - - ## 3 < 2e-16 0.055 - ## 4 < 2e-16 3.1e-10 1.8e-14 ## ## P value adjustment method: holm ``` --- class: inverse Assumptions and diagnostics ## Next time... <!-- ## Contrasts --> <!-- You can change which group is the reference: --> <!-- ```{r```{r} --> <!-- contrasts(rotate$Group) = contr.treatment(n = 4, base = 3) --> <!-- coef(summary(lm(Time~Group, data = rotate))) --> <!-- ```} --> <!-- contrasts(rotate$Group) = contr.treatment(n = 4, base = 3) --> <!-- coef(summary(lm(Time~Group, data = rotate))) %>% kable(., digits = c(2,2,2,3)) %>% kable_styling() --> <!-- ``` --> <!-- You're not restricted to simple dummy coding. Choosing another variant of contrast codes allows you to test more specific hypotheses. There are some common coding schemes. --> <!-- --- --> <!-- For example, **deviation coding** (sometimes called **"effect coding"**): --> <!-- ```{r} --> <!-- contr.sum(4) --> <!-- ``` --> <!-- ```{r, results='asis'} --> <!-- contrasts(rotate$Group) = contr.sum(n = 4) --> <!-- coef(summary(lm(Time~Group, data = rotate))) %>% kable(., digits = c(2,2,2,3)) %>% kable_styling() --> <!-- ``` --> <!-- --- --> <!-- ### Contrasts --> <!-- You can create a contrast matrix that tests any number of hypotheses, like whether groups 1 and 3 are different from group 4. The rules for setting up contrast codes are: --> <!-- 1. The sum of the weights across all groups for each code variable must equal 0. (Sum down the column = 0). --> <!-- 2. The sum of the product of each pair of code variables, `\(C_1C_2,\)` must equal 0. (This is challenging when your groups are unequal sizes). --> <!-- 3. The difference between the value of the set of positive weights and the set of negative weights should equal 1 for each code variable (column). --> <!-- One benefit* to using the ANOVA framework instead of the regression framework is that you can estimate the total variability captured by one categorical variable controlling for another categorical or continuous variable with ease. --> <!-- `\(\eta^2\)` (eta-squared) is a standardized measure of effect size used in analyses of variance. This effect size is the **proportion of variance in Y that is accounted for by one independent variable.** --> <!-- `$$\eta^2 = \frac{SS_{variable}}{SS_{total}}$$` --> <!-- For example, what if we believed gender was associated with performance on the noise burst task. --> <!-- .small[* No difference when using R though.] --> <!-- --- --> <!-- ```{r, echo = F} --> <!-- set.seed(200620) --> <!-- hightime = sample(x = c("Male", "Female"), size = nrow(rotate), replace = T, prob = c(.75,.25)) --> <!-- lowtime = sample(x = c("Male", "Female"), size = nrow(rotate), replace = T, prob = c(.25,.75)) --> <!-- rotate$gender = hightime --> <!-- rotate$gender[rotate$Time < 425] = lowtime[rotate$Time < 425] --> <!-- ``` --> <!-- ```{r} --> <!-- mod2 = aov(Time ~ Group + gender, data = rotate) --> <!-- anova(mod2) --> <!-- ``` -->