class: center, middle, inverse, title-slide # Interactions (IV) --- ## Last time... - Factorial ANOVA --- .pull-left[ The example data are from a simulated study in which 180 participants performed an eye-hand coordination task in which they were required to keep a mouse pointer on a red dot that moved in a circular motion. ] .pull-right[  ] The outcome was the time of the 10th failure. The experiment used a completely crossed, 3 x 3 factorial design. One factor was dot speed: .5, 1, or 1.5 revolutions per second. The second factor was noise condition: no noise, controllable noise, and uncontrollable noise. The design was balanced. --- ### Marginal/cell means <table class="table" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Noise </th> <th style="text-align:right;"> Slow </th> <th style="text-align:right;"> Medium </th> <th style="text-align:right;"> Fast </th> <th style="text-align:right;"> Marginal </th> </tr> </thead> <tbody> <tr grouplength="3"><td colspan="5" style="border-bottom: 1px solid;"><strong></strong></td></tr> <tr> <td style="text-align:left; padding-left: 2em;" indentlevel="1"> None </td> <td style="text-align:right;background-color: #EECACA !important;"> 630.72 </td> <td style="text-align:right;background-color: #B2D4EB !important;"> 525.29 </td> <td style="text-align:right;background-color: #FFFFC5 !important;"> 329.28 </td> <td style="text-align:right;color: white !important;background-color: grey !important;"> 495.10 </td> </tr> <tr> <td style="text-align:left; padding-left: 2em;" indentlevel="1"> Controllable </td> <td style="text-align:right;background-color: #EECACA !important;"> 576.67 </td> <td style="text-align:right;background-color: #B2D4EB !important;"> 492.72 </td> <td style="text-align:right;background-color: #FFFFC5 !important;"> 287.23 </td> <td style="text-align:right;color: white !important;background-color: grey !important;"> 452.21 </td> </tr> <tr> <td style="text-align:left; padding-left: 2em;" indentlevel="1"> Uncontrollable </td> <td style="text-align:right;background-color: #EECACA !important;"> 594.44 </td> <td style="text-align:right;background-color: #B2D4EB !important;"> 304.62 </td> <td style="text-align:right;background-color: #FFFFC5 !important;"> 268.16 </td> <td style="text-align:right;color: white !important;background-color: grey !important;"> 389.08 </td> </tr> <tr> <td style="text-align:left;background-color: white !important;"> Marginal </td> <td style="text-align:right;background-color: #EECACA !important;background-color: white !important;"> 600.61 </td> <td style="text-align:right;background-color: #B2D4EB !important;background-color: white !important;"> 440.88 </td> <td style="text-align:right;background-color: #FFFFC5 !important;background-color: white !important;"> 294.89 </td> <td style="text-align:right;color: white !important;background-color: grey !important;background-color: white !important;"> 445.46 </td> </tr> </tbody> </table> --- ## Running the analysis of variance ```r fit = lm(Time ~ Speed*Noise, data = Data) anova(fit) ``` ``` ## Analysis of Variance Table ## ## Response: Time ## Df Sum Sq Mean Sq F value Pr(>F) ## Speed 2 2805871 1402936 109.3975 < 2.2e-16 *** ## Noise 2 341315 170658 13.3075 4.252e-06 *** ## Speed:Noise 4 295720 73930 5.7649 0.0002241 *** ## Residuals 171 2192939 12824 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` --- ```r lsr::etaSquared(fit) ``` ``` ## eta.sq eta.sq.part ## Speed 0.49786168 0.5613078 ## Noise 0.06056150 0.1346807 ## Speed:Noise 0.05247123 0.1188269 ``` An effect size, `\(\eta^2\)`, provides a simple way of indexing effect magnitude for ANOVA designs, especially as they get more complex. `\(\eta^2\)` represents the proportion of variance in Y explained by a single factor (or interaction of factors). It is identical in its calculation to `\(R^2\)`: `$$\large \eta^2 = \frac{SS_{\text{effect}}}{SS_{\text{Y}}}$$` --- ```r lsr::etaSquared(fit) ``` ``` ## eta.sq eta.sq.part ## Speed 0.49786168 0.5613078 ## Noise 0.06056150 0.1346807 ## Speed:Noise 0.05247123 0.1188269 ``` In an experimental design, variance in Y is created, rather than observed, by manipulating participants. The ways in which an experimenter chooses to manipulate particiapnts can increase or decrease variance in Y, making it difficult to compare the effect of a single manipulation across studies with different designs. Partial `\(\eta^2\)` is meant to aid interpretation by considering only the variance associated with an effect and random variablity, which is naturally occuring and not under the control of the experimenter. `$$\large \text{Partial }\eta^2 = \frac{SS_{\text{effect}}}{SS_{\text{effect}} + SS_{\text{within}}}$$` --- ### Differences in means In a factorial design, marginal means or cell means must be calculated in order to interpret main effects and the interaction, respectively. The confidence intervals around those means likewise are needed. <table class="table" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Noise </th> <th style="text-align:right;"> Slow </th> <th style="text-align:right;"> Medium </th> <th style="text-align:right;"> Fast </th> <th style="text-align:right;"> Marginal </th> </tr> </thead> <tbody> <tr grouplength="3"><td colspan="5" style="border-bottom: 1px solid;"><strong></strong></td></tr> <tr> <td style="text-align:left; padding-left: 2em;" indentlevel="1"> None </td> <td style="text-align:right;"> 630.72 </td> <td style="text-align:right;"> 525.29 </td> <td style="text-align:right;"> 329.28 </td> <td style="text-align:right;"> 495.10 </td> </tr> <tr> <td style="text-align:left; padding-left: 2em;" indentlevel="1"> Controllable </td> <td style="text-align:right;"> 576.67 </td> <td style="text-align:right;"> 492.72 </td> <td style="text-align:right;"> 287.23 </td> <td style="text-align:right;"> 452.21 </td> </tr> <tr> <td style="text-align:left; padding-left: 2em;" indentlevel="1"> Uncontrollable </td> <td style="text-align:right;"> 594.44 </td> <td style="text-align:right;"> 304.62 </td> <td style="text-align:right;"> 268.16 </td> <td style="text-align:right;"> 389.08 </td> </tr> <tr> <td style="text-align:left;"> Marginal </td> <td style="text-align:right;"> 600.61 </td> <td style="text-align:right;"> 440.88 </td> <td style="text-align:right;"> 294.89 </td> <td style="text-align:right;"> 445.46 </td> </tr> </tbody> </table> These means will be based on different sample sizes, which has an impact on the width of the confidence interval. --- ### Precision If the homogeneity of variances assumption holds, a common estimate of score variability `\((MS_{within})\)` underlies all of the confidence intervals. `$$\large SE_{mean} = \sqrt{\frac{MS_{within}}{N}}$$` `$$\large CI_{mean} = Mean \pm t_{df_{within}, \alpha = .05}\sqrt{\frac{MS_{within}}{N}}$$` The sample size, `\(N\)`, depends on how many cases are aggregated to create the mean. The `\(MS_{within}\)` is common to all calculations if homogeneity of variances is met. The degrees of freedom for `\(MS_{within}\)` determine the value of `\(t\)` to use. --- ```r anova(fit) ``` ``` ## Analysis of Variance Table ## ## Response: Time ## Df Sum Sq Mean Sq F value Pr(>F) ## Speed 2 2805871 1402936 109.3975 < 2.2e-16 *** ## Noise 2 341315 170658 13.3075 4.252e-06 *** ## Speed:Noise 4 295720 73930 5.7649 0.0002241 *** ## Residuals 171 2192939 12824 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` --- ```r library(emmeans) (time_rg = ref_grid(fit)) ``` ``` ## 'emmGrid' object with variables: ## Speed = Slow, Medium, Fast ## Noise = None, Controllable, Uncontrollable ``` ```r summary(time_rg) ``` ``` ## Speed Noise prediction SE df ## Slow None 631 25.3 171 ## Medium None 525 25.3 171 ## Fast None 329 25.3 171 ## Slow Controllable 577 25.3 171 ## Medium Controllable 493 25.3 171 ## Fast Controllable 287 25.3 171 ## Slow Uncontrollable 594 25.3 171 ## Medium Uncontrollable 305 25.3 171 ## Fast Uncontrollable 268 25.3 171 ``` --- The lsmeans( ) function produces marginal and cell means along with their confidence intervals. These are the marginal means for the Noise main effect. ```r noise_lsm = emmeans::lsmeans(time_rg, "Noise") ``` ``` ## NOTE: Results may be misleading due to involvement in interactions ``` ```r noise_lsm ``` ``` ## Noise lsmean SE df lower.CL upper.CL ## None 495 14.6 171 466 524 ## Controllable 452 14.6 171 423 481 ## Uncontrollable 389 14.6 171 360 418 ## ## Results are averaged over the levels of: Speed ## Confidence level used: 0.95 ``` --- ```r library(sjPlot) noise_m = plot_model(fit, type = "emm", terms = c("Noise")) + theme_sjplot(base_size = 20) speed_m = plot_model(fit, type = "emm", terms = c("Speed")) + theme_sjplot(base_size = 20) library(ggpubr) ggarrange(noise_m, speed_m, ncol = 2) ``` 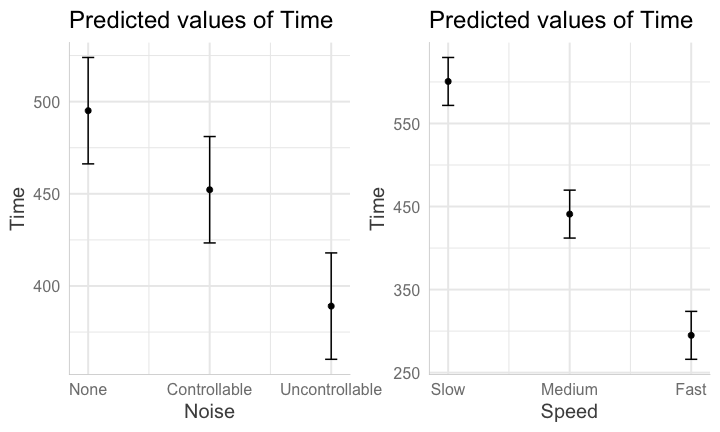<!-- --> --- ```r plot_model(fit, type = "pred", terms = c("Speed", "Noise")) + theme_sjplot(base_size = 20) + theme(legend.position = "bottom") ``` 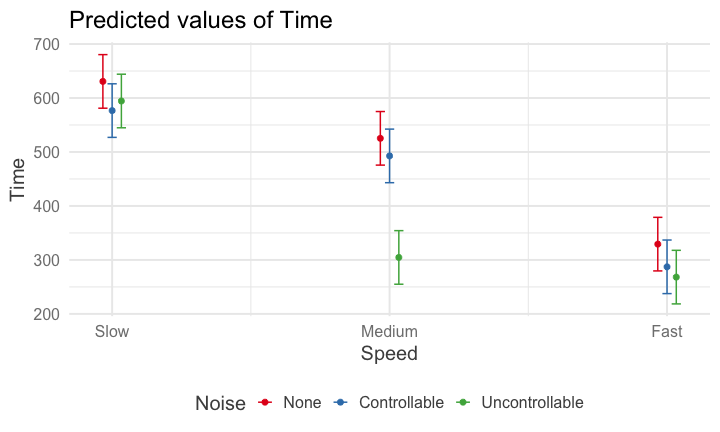<!-- --> --- ### Precision A reminder that comparing the confidence intervals for two means (overlap) is not the same as the confidence interval for the difference between two means. $$ `\begin{aligned} \large SE_{\text{mean}} &= \sqrt{\frac{MS_{within}}{N}}\\ \large SE_{\text{mean difference}} &= \sqrt{MS_{within}[\frac{1}{N_1}+\frac{1}{N_2}]} \\ \large SE_{\text{mean difference}} &= \sqrt{\frac{2MS_{within}}{N}} \\ \end{aligned}` $$ --- ### Cohen's D `\(\eta^2\)` is useful for comparing the relative effect sizes of one factor to another. If you want to compare the differences between groups, Cohen's d is the more appropriate metric. Like in a t-test, you'll divide the differences in means by the pooled standard deviation. The pooled variance estimate is the `\(MS_{error}\)` ```r fit = lm(Time ~ Speed*Noise, data = Data) anova(fit)[,"Mean Sq"] ``` ``` ## [1] 1402935.71 170657.62 73929.93 12824.20 ``` ```r MS_error = anova(fit)[,"Mean Sq"][4] ``` So to get the pooled standard deviation: ```r pooled_sd = sqrt(MS_error) ``` --- ### Cohen's D ```r (noise_df = as.data.frame(noise_lsm)) ``` ``` ## Noise lsmean SE df lower.CL upper.CL ## 1 None 495.0976 14.61974 171 466.2392 523.9560 ## 2 Controllable 452.2079 14.61974 171 423.3495 481.0663 ## 3 Uncontrollable 389.0759 14.61974 171 360.2175 417.9343 ``` ```r (d_none_control = diff(noise_df[c(1,2), "lsmean"])/pooled_sd) ``` ``` ## [1] -0.3787367 ``` --- ### Follow-up comparisons Interpretation of the main effects and interaction in a factorial design will usually require follow-up comparisons. These need to be conducted at the level of the effect. Interpretation of a main effect requires comparisons among the marginal means. Interpretation of the interaction requires comparisons among the cell means. The `emmeans` package makes these comparisons very easy to conduct. --- ```r noise_lsm = emmeans::lsmeans(time_rg, "Noise") ``` ``` ## NOTE: Results may be misleading due to involvement in interactions ``` ```r pairs(noise_lsm, adjust = "holm") ``` ``` ## contrast estimate SE df t.ratio p.value ## None - Controllable 42.9 20.7 171 2.074 0.0395 ## None - Uncontrollable 106.0 20.7 171 5.128 <.0001 ## Controllable - Uncontrollable 63.1 20.7 171 3.053 0.0052 ## ## Results are averaged over the levels of: Speed ## P value adjustment: holm method for 3 tests ``` --- ## Assumptions You can check the assumptions of the factorial ANOVA in much the same way you check them for multiple regression; but given the categorical nature of the predictors, some assumptions are easier to check. Homogeneity of variance, for example, can be tested using Levene's test, instead of examining a plot. ```r library(car) leveneTest(Time ~ Speed*Noise, data = Data) ``` ``` ## Levene's Test for Homogeneity of Variance (center = median) ## Df F value Pr(>F) ## group 8 0.5879 0.787 ## 171 ``` --- ## Unbalanced designs If designs are balanced, then the main effects and interpretation effects are independent/orthogonal. In other words, knowing what condition a case is in on Variable 1 will not make it any easier to guess what condition they were part of in Variable 2. However, if your design is unbalanced, the main effects and interaction effect are partly confounded. ```r table(Data2$Level, Data2$Noise) ``` ``` ## ## Controllable Uncontrollable ## Soft 10 30 ## Loud 20 20 ``` --- ### Sums of Squares .pull-left[ In a Venn diagram, the overlap represents the variance shared by the variables. Now there is variance accounted for in the outcome (Time) that cannot be unambiguously attributed to just one of the predictors There are several options for handling the ambiguous regions.] .pull-right[ 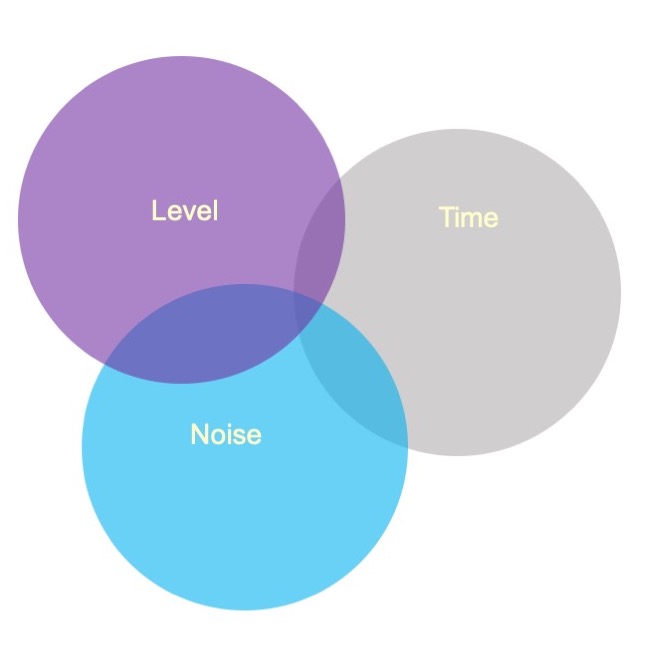 ] --- ### Sums of Squares (III) .pull-left[ ] .pull-right[ There are three basic ways that overlapping variance accounted for in the outcome can be handled. These are known as Type I, Type II, and Type III sums of squares. Type III sums of squares is the easiest to understand—the overlapping variance accounted for goes unclaimed by any variable. Effects can only account for the unique variance that they share with the outcome. ] --- ### Sums of Squares (I) .pull-left[ Type I sums of squares allocates the overlapping variance according to some priority rule. All of the variance is claimed, but the rule needs to be justified well or else suspicions about p-hacking are likely. The rule might be based on theory or causal priority or methodological argument. ] .pull-right[ ] --- ### Sums of Squares (I) If a design is quite unbalanced, different orders of effects can produce quite different results. ```r fit_1 = aov(Time ~ Noise + Level, data = Data2) summary(fit_1) ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## Noise 1 579478 579478 41.78 8.44e-09 *** ## Level 1 722588 722588 52.10 3.20e-10 *** ## Residuals 77 1067848 13868 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ```r lsr::etaSquared(fit_1, type = 1) ``` ``` ## eta.sq eta.sq.part ## Noise 0.2445144 0.3517689 ## Level 0.3049006 0.4035822 ``` --- ### Sums of Squares (I) If a design is quite unbalanced, different orders of effects can produce quite different results. ```r fit_1 = aov(Time ~ Level + Noise, data = Data2) summary(fit_1) ``` ``` ## Df Sum Sq Mean Sq F value Pr(>F) ## Level 1 1035872 1035872 74.69 5.86e-13 *** ## Noise 1 266194 266194 19.20 3.68e-05 *** ## Residuals 77 1067848 13868 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ```r lsr::etaSquared(fit_1, type = 1) ``` ``` ## eta.sq eta.sq.part ## Level 0.4370927 0.4924002 ## Noise 0.1123222 0.1995395 ``` --- ### Sums of Squares (II) Type II sums of squares are the most commonly used but also a bit more complex to understand. An approximately true description is that an effect is allocated the proportion of variance in the outcome that is unshared with effects at a similar or lower level. A technically correct description is that a target effect is allocated the proportion of variance in the outcome that is unshared with other effects that do not contain the target. --- ### Sums of Squares (II) In an A x B design, the A effect is the proportion of variance that is accounted for by A after removing the variance accounted for by B. The A x B interaction is allocated the proportion of variance that is accounted for by A x B after removing the variance accounted for by A and B. There is no convenient way to illustrate this in a Venn diagram. --- ### Sums of Squares Let R(·) represent the residual sum of squares for a model, so for example R(A,B,AB) is the residual sum of squares fitting the whole model, R(A) is the residual sum of squares fitting just the main effect of A, and R(1) is the residual sum of squares fitting just the mean. | Effect | Type I | Type II | Type III | |--------|-------------------|-------------------|---------------------| | A | R(1)-R(A) | R(B)-R(A,B) | R(B,A:B)-R(A,B,A:B) | | B | R(A)-R(A,B) | R(A)-R(A,B) | R(A,A:B)-R(A,B,A:B) | | A:B | R(A,B)-R(A,B,A:B) | R(A,B)-R(A,B,A:B) | R(A,B)-R(A,B,A:B) | --- ### Sums of Squares Let R(·) represent the residual sum of squares for a model, so for example R(A,B,AB) is the residual sum of squares fitting the whole model, R(A) is the residual sum of squares fitting just the main effect of A, and R(1) is the residual sum of squares fitting just the mean. | Effect | Type I | Type II | Type III | |--------|-------------------|-------------------|---------------------| | A | SS(A) | SS(A|B) | SS(A|B,AB) | | B | SS(B|A) | SS(B|A) | SS(B|A,AB) | | A:B | SS(AB|A,B) | SS(AB|A,B) | SS(AB|A,B) | --- The `aov( )` function in R produces Type I sums of squares. The `Anova( )` function from the car package provides Type II and Type III sums of squares. These work as expected provided the predictors are factors. ```r Anova(fit, type = "II") ``` ``` ## Anova Table (Type II tests) ## ## Response: Time ## Sum Sq Df F value Pr(>F) ## Speed 2805871 2 109.3975 < 2.2e-16 *** ## Noise 341315 2 13.3075 4.252e-06 *** ## Speed:Noise 295720 4 5.7649 0.0002241 *** ## Residuals 2192939 171 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` --- All of the between-subjects variance is accounted for by an effect in Type I sums of squares. The sums of squares for each effect and the residual will equal the total sum of squares. For Type II and Type III sums of squares, the sums of squares for effects and residual will be less than the total sum of squares. Some variance (in the form of SS) goes unclaimed. The highest order effect (assuming standard ordering) has the same SS in all three models. When a design is balanced, Type I, II, and III sums of squares are equivalent. --- ## Summary (and Regression) Factorial ANOVA is the method by which we can examine whether two (or more) categorical IVs have joint effects on a continuous outcome of interest. Like all general linear models, factorial ANOVA is a specific case of multiple regression. However, we may choose to use an ANOVA framework for the sake of interpretability. --- .pull-left[ #### Factorial ANOVA Interaction tests whether there are differences in differences. A main effect is the effect of one IV on the DV **ignoring the other variable(s)**. ] .pull-right[ #### Regression Interaction tests whether slope changes. A conditional effect is the effect of IV on the DV **assuming all the other variables are 0**. ] --- ## Power What is (statistical) power? How can we increase power? -- The likelihood of finding an effect *if the effect actually exists.* Power gets larger as we: * increase our sample size * reduce (error) variance * raise our Type I error rate * study larger effects --- <!-- --> --- ## Power in multiple regression (additive effects) When calculating power for the omnibus test, use the expected multiple `\(R^2\)` value to calculate an effect size: `$$\large f^2 = \frac{R^2}{1-R^2}$$` --- ### Omnibus power ```r R2 = .10 (f = R2/(1-R2)) ``` ``` ## [1] 0.1111111 ``` ```r library(pwr) pwr.f2.test(u = 3, # number of predictors in the model f2 = f, sig.level = .05, #alpha power =.90) # desired power ``` ``` ## ## Multiple regression power calculation ## ## u = 3 ## v = 127.5235 ## f2 = 0.1111111 ## sig.level = 0.05 ## power = 0.9 ``` `v` is the denominator df of freedom, so the number of participants needed is v + p + 1. --- ### Coefficient power To estimate power for a single coefficient, you need to consider (1) how much variance is accounted for by just the variable and (2) how much variance you'll account for in Y overall. `$$\large f^2 = \frac{R^2_Y-R^2_{Y.X}}{1-R_Y^2}$$` --- ### Coefficient power ```r R2 = .10 RX1 = .03 (f = (R2-RX1)/(1-R2)) ``` ``` ## [1] 0.07777778 ``` ```r pwr.f2.test(u = 3, # number of predictors in the model f2 = f, sig.level = .05, #alpha power =.90) # desired power ``` ``` ## ## Multiple regression power calculation ## ## u = 3 ## v = 182.1634 ## f2 = 0.07777778 ## sig.level = 0.05 ## power = 0.9 ``` `v` is the denominator df of freedom, so the number of participants needed is v + p + 1. --- ## Effect sizes (interactions) To start our discussion on powering interaction terms, we need to first consider the effect size of an interaction. How big can we reasonably expect an interaction to be? - Interactions are always partialled effects; that is, we examine the relationship between the product of variables X and Z only after we have controlled for X and controlled for Z. How does this affect the size of the relationship between XZ and Y? ??? The effect of XZ and Y will be made smaller as X or Z (or both) is related to the product -- the semi-partial correlation is always smaller than or equal to the zero-order correlation. --- ## McClelland and Judd (1993) Is it more difficult to find interaction effects in experimental studies or observational studies? -- What factors make it relatively easier to find interactions in experimental work? --- ### Factors influencing power in experimental studies - No measurement error of IV * don't have to guess what condition a participant is in * measurement error is exacerbated when two variables measured with error are multiplied by each other - Experimentalists can force cross-over interactions; observational studies may be restricted to fan interactions * cross-over interactions are easier to detect than fan interactions - Experimentalists can concentrate scores on extreme ends on both X and Z * in observational studies, data tends to cluster around the mean * increases variability in both X and Z, and in XZ - Experimentalists can also force orthognality in X and Z ??? Other things: you can insure that you study the full range of X in an experiment, but you may have restricted range in an observational study --- ### McClelland and Judd's simulation For the experiment simulations, we used 2 X 2 factorial designs, with values of X and Z equal to +1 and —1 and an equal number of observations at each of the four combinations of X and Z values. ```r X = rep(c(-1,1), each = 50) Z = rep(c(-1,1), times = 50) table(X,Z) ``` ``` ## Z ## X -1 1 ## -1 25 25 ## 1 25 25 ``` --- ### McClelland and Judd's simulation For the field study simulations, we used values of X and Z that varied between the extreme values of +1 and —1. More specifically, in the field study simulations, values of X and Z were each sampled independently from a normal distribution with a mean of 0 and a standard deviation of 0.5. Values of X and Z were rounded to create equally spaced 9-point scales ranging from -1 to +1 because ranges in field studies are always finite and because ratings are often on scales with discrete intervals. ```r X = rnorm(n = 100, mean = 0, sd = .5) Z = rnorm(n = 100, mean = 0, sd = .5) X = round(X/.2)*.2 Z = round(Z/.2)*.2 psych::describe(data.frame(X,Z), fast = T) ``` ``` ## vars n mean sd min max range se ## X 1 100 0.02 0.47 -1.2 1.0 2.2 0.05 ## Z 2 100 -0.05 0.50 -1.2 1.2 2.4 0.05 ``` --- For the simulations of both the field studies and the experiments, `\(\beta_0 = 0, \beta_X=\beta_Z=\beta_{XZ} = 1.\)` There were 100 observations, and errors for the model were sampled from the same normal distribution with a mean of 0 and a standard deviation of 4. ```r Y = 0 + 1*X + 1*Z + 1*X*Z + rnorm(n = 100, mean = 0, sd = 4) summary(lm(Y ~ X*Z)) ``` ``` ## ## Call: ## lm(formula = Y ~ X * Z) ## ## Residuals: ## Min 1Q Median 3Q Max ## -12.2691 -1.8418 -0.0396 2.6666 9.1478 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 0.1573 0.4070 0.386 0.700 ## X 0.7812 0.8712 0.897 0.372 ## Z 0.6686 0.8108 0.825 0.412 ## X:Z 0.9635 1.9267 0.500 0.618 ## ## Residual standard error: 4.043 on 96 degrees of freedom ## Multiple R-squared: 0.01752, Adjusted R-squared: -0.01318 ## F-statistic: 0.5706 on 3 and 96 DF, p-value: 0.6357 ``` --- From 100 simulations each, estimates of the model parameter `\(\beta_{XZ}\)` the moderator or interaction effect equaled 0.977 and 0.979 for the field studies and experiments, respectively. ```r set.seed(0305) ``` .pull-left[ ```r # for experimental studies sim = 100 ebeta_xz = numeric(length = 100) et_xz = numeric(length = 100) for(i in 1:sim){ # simulate data X = rep(c(-1,1), each = 50) Z = rep(c(-1,1), times = 50) Y = 0 + 1*X + 1*Z + 1*X*Z + rnorm(n = 100, mean = 0, sd = 4) #run model model = lm(Y ~ X*Z) coef = coef(summary(model)) #extract coefficients beta = coef["X:Z", "Estimate"] t_val = coef["X:Z", "t value"] #save to vectors ebeta_xz[i] = beta et_xz[i] = t_val } ``` ] .pull-right[ ```r # for observational studies obeta_xz = numeric(length = 100) ot_xz = numeric(length = 100) for(i in 1:sim){ # simulate data X = rnorm(n = 100, mean=0, sd = .5) Z = rnorm(n = 100, mean=0, sd = .5) X = round(X/.2)*.2 Z = round(Z/.2)*.2 Y = 0 + 1*X + 1*Z + 1*X*Z + rnorm(n = 100, mean = 0, sd = 4) #run model model = lm(Y ~ X*Z) coef = coef(summary(model)) #extract coefficients beta = coef["X:Z", "Estimate"] t_val = coef["X:Z", "t value"] #save to vectors obeta_xz[i] = beta ot_xz[i] = t_val } ``` ] --- ```r mean(ebeta_xz) ``` ``` ## [1] 0.9440304 ``` ```r mean(obeta_xz) ``` ``` ## [1] 1.175444 ``` <!-- --> --- ```r mean(et_xz) ``` ``` ## [1] 2.383435 ``` ```r mean(ot_xz) ``` ``` ## [1] 0.7411209 ``` <!-- --> --- ```r cv = qt(p = .975, df = 100-3-1) esig = et_xz > cv sum(esig) ``` ``` ## [1] 66 ``` ```r osig = ot_xz > cv sum(osig) ``` ``` ## [1] 12 ``` In our simulation, 66% of experimental studies were statistically significant, whereas only 12% of observational studies were significant. Remember, we built our simulation based on data where there really is an interaction effect (i.e., the null is false). McClelland and Judd found that 74% of experimental studies and 9% of observational studies were significant. --- ### Efficiency <img src="images/efficiency.png" width="55%" /> ??? Efficiency = the ratio of the variance of XZ (controlling for X and Z) of a design to the best possible design (upper right corner). High efficiency is better; best efficiency is 1. --- ### Efficiency .pull-left[ If the optimal design has N obserations, then to have the same standard error (i.e., the same power), any other design needs to have N*(1/efficency). So a design with .06 efficency needs `\(\frac{1}{.06} = 16.67\)` times the sample size to detect the effect. ] .pull-right[  ] This particular point has been ["rediscovered"](https://statmodeling.stat.columbia.edu/2018/03/15/need-16-times-sample-size-estimate-interaction-estimate-main-effect/) as recently as 2018: * you need 16 times the sample size to detect an interaction as you need for a main effect of the same size. ??? This generalizes to higher-order interactions as well. If you have a three-way interaction, you need 16*16 (256 times the number of people). --- ## Observational studies: What NOT to do Recode X and Z into more extreme values (e.g., median splits) * while this increases variance in X and Z, it also increases measurement error Collect a random sample and then only perform analyses on the subsample with extreme values * reduces sample size and also generalizability #### What can be done? M&J suggest oversampling extremes and using weighted and unweighted samples --- ## Experimental studies: What NOT to do Be mean to field researchers Forget about lack of external validity and generalizability Ignore power when comparing interaction between covariate and experimental predictors (ANCOVA or multiple regression with categorical and continuous predictors)